






 |  |  |  |  |  | 
|
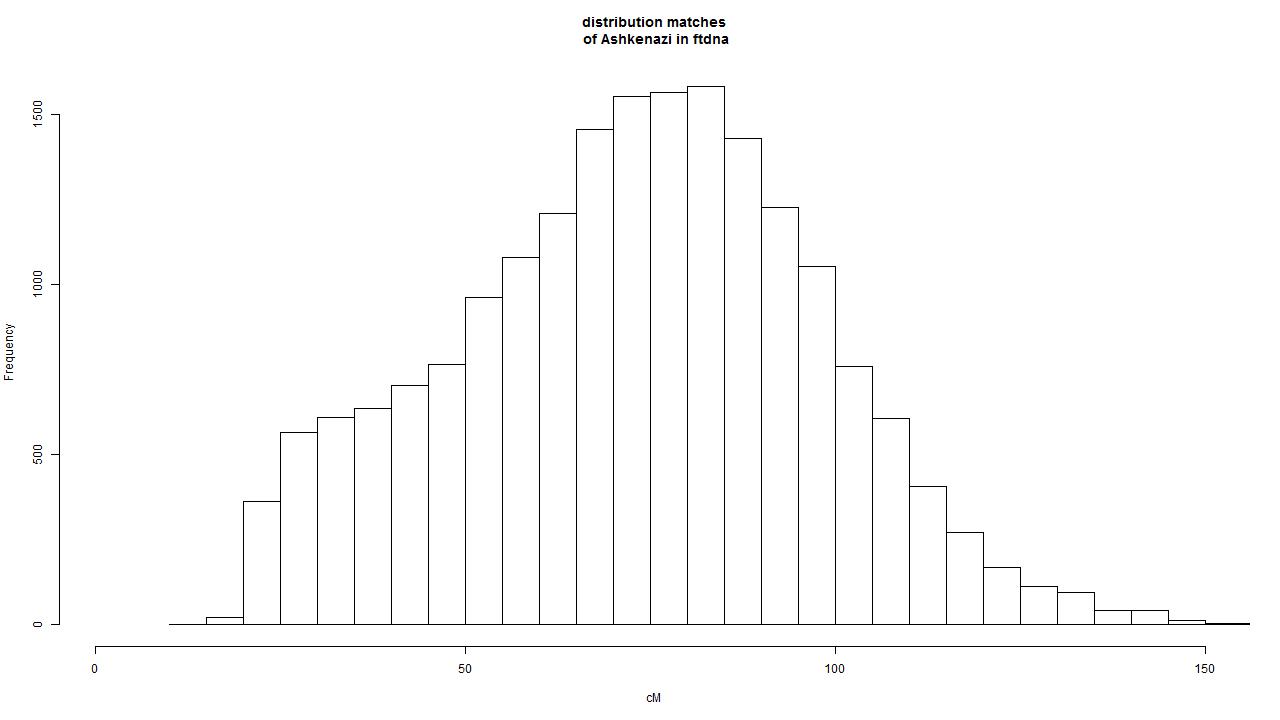
I selected 32 non-related persons that have 100% Ashkenazi descendancy and listed the matches of these 32 persons. This resulted in a grand total of 468402 matches. I selected the persons that have at least three matches above 80cM. This resulted in 19603 unique persons. In the next step the people were selected that have a mtDNA branch that is only reported if mtDNAfullsequence test is done. This resulted in 3589 mtDNAfullsequence branch results. These are the results that are used in the table below.
Additional the public results of the mtDNA phylogenetic tree of ftdna was used to determine the number of people that were measured by ftdna. This was determined as the sum of the people in the first 15 countries. Since I don't have the Unknown country, I miss 32%, which is the average of the complete sample. In the extreme case of a large branch (e.g. 1212 persons in H1) I miss 10% in the countries that are outside the first 15 countries. This does not change the pattern that I find.
mtdna is relatively small and the number of mutations in a given timeperiod is small. The average time for a mutation is about 1/16.569*2.5*10^-8 which is about 2400 years. This means that I will have four types of Jewish mtdna branchs:
Only six of the 90 branches with six 100% Ashkenazi persons had a branch with both clear Ashkenazi and non-Jewish people. Given the mutation rate of 2400 years for mtdna, 7% is quite small. This was the case for the branches H1b1-T16362C +5899.1C+A8348G , H6a1a3 +A16482G+C16519T, H11a1 +C16519T, H56, H65a, K2a. Given the history of Ashkenazi, the distribution of mtdna haplogroups and the size of these branches, it seems likely that a significant part of these six branches descent from a European ancestor that became Jewish.
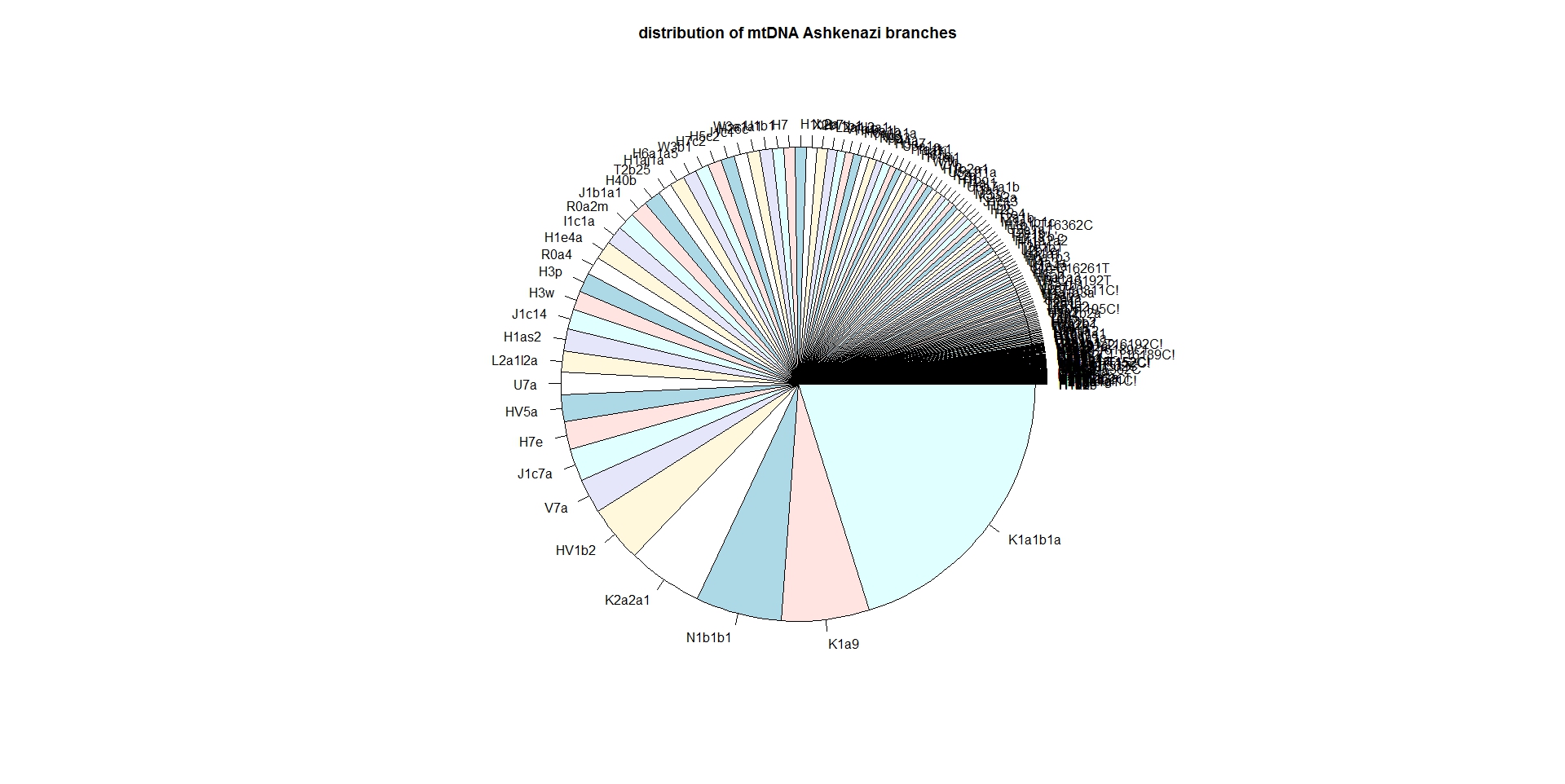
If you click on "# of persons", that column is sorted; if you click twice on "# of persons" the largest branch is shown first.
The reference to yfull and ftdna can be interesting. The ftdna data has more samples; the yfull data has from scientific publications and shows subbranches. Notice that the amount of registered samples is the result of a collection of people who were tested. They do not represent a representative collection of data.
| Branch # | mtdna name | approximate | SNPs of Jewish branch or comment | # of persons | # in ftdna branch | ybp of Behar, van Oven et al. (2012) |
| MB-001 | A-T152C!-T16189C! | yfull ftdna | 2 | 4 | 0+-0 | |
| MB-002 | H1a3a | yfull ftdna | 2 | 96 | 3850+-1620 | |
| MB-003 | H1a3b | yfull ftdna | 1 | 74 | 3150+-2060 | |
| MB-004 | H1b | yfull ftdna | G15172A | 9 | 259 | 6230+-1800 |
| MB-005 | H1b1-T16362C | yfull ftdna | non-Ashkenazi in SNP-group 5899.1C+A8348G | 11 | 408 | 0+-0 |
| MB-006 | H1b2 | yfull ftdna | 1 | 67 | 3660+-2770 | |
| MB-007 | H1b2a+H1b2a1 | yfull ftdna | 42 | 49 | 700+-4060 | |
| MB-009 | H1b5 | yfull ftdna | 1 | 17 | 0+-0 | |
| MB-010 | H1f | yfull ftdna | T7309C+C8409T+T12945C+G15221A+A16183c | 3 | 38 | 3670+-3620 |
| MB-011 | H1c3 | yfull ftdna | 1 | 215 | 4530+-1850 | |
| MB-012 | H1e4+H1e4a | yfull ftdna | - | 57 | 84 | 2730+-7140 |
| MB-014 | H1o | yfull ftdna | 1 | 39 | 2480+-3250 | |
| MB-015 | H1u2 | yfull ftdna | insufficient access | 4 | 13 | 0+-0 |
| MB-016 | H1ai1 | yfull ftdna | - | 13 | 24 | 0+-0 |
| MB-017 | H1aj | yfull ftdna | insufficient access | 6 | 21 | 8170+-2950 |
| MB-018 | H1aj1+H1aj1a | yfull ftdna | 50 | 54 | 0+-0 | |
| MB-020 | H1as2 | yfull ftdna | 52 | 51 | 1650+-2230 | |
| MB-021 | H1ax | yfull ftdna | 2 | 15 | 4940+-2860 | |
| MB-022 | H1bd | yfull ftdna | 1 | 18 | 0+-0 | |
| MB-023 | H1bo | yfull ftdna | - | 13 | 24 | 4480+-2170 |
| MB-024 | H1bw | yfull ftdna | insufficient access | 3 | 26 | 6110+-3690 |
| MB-025 | H2a2a1g | yfull ftdna | 1 | 18 | 0+-0 | |
| MB-026 | H2a2b1 | yfull ftdna | T16224C+A16235G+C16519T | 4 | 80 | 4390+-2080 |
| MB-027 | H2a2b1a1 | yfull ftdna | 1 | 68 | 1810+-1630 | |
| MB-028 | H3 | yfull ftdna | C10655T+T16298C+A16497G | 8 | 386 | 8910+-1060 |
| MB-029 | H3k1a | yfull ftdna | 1 | 70 | 0+-0 | |
| MB-030 | H3-T16311C! | yfull ftdna | 1 | 57 | 0+-0 | |
| MB-031 | H3p | yfull ftdna | 46 | 60 | 6370+-2910 | |
| MB-032 | H3w | yfull ftdna | 47 | 54 | 3230+-2310 | |
| MB-033 | H3ap | yfull ftdna | 2 | 63 | 5700+-3770 | |
| MB-034 | H4a1a1 | yfull ftdna | 1 | 165 | 6600+-2080 | |
| MB-035 | H4a1a1a | yfull ftdna | C16519T+ C152T! | 3 | 268 | 5890+-1790 |
| MB-036 | H4a1a3a | yfull ftdna | G513A+A14133G+C16519T | 6 | 19 | 2370+-2400 |
| MB-037 | H5'36 | yfull ftdna | 2 | 33 | 0+-0 | |
| MB-038 | H5 | yfull ftdna | insufficient access | 3 | 147 | 9870+-1400 |
| MB-039 | H5a | yfull ftdna | 1 | 19 | 9110+-1380 | |
| MB-040 | H5a1 | yfull ftdna | non-Ashkenazi in SNP-group C16519T | 5 | 474 | 6560+-1530 |
| MB-041 | H5a-T152C! | yfull ftdna | 1 | 20 | 0+-0 | |
| MB-042 | H5a7 | yfull ftdna | C16519T | 18 | 40 | 1510+-2050 |
| MB-043 | H5c2 | yfull ftdna | 33 | 37 | 0+-0 | |
| MB-044 | H5-C16192T | yfull ftdna | insufficient access | 7 | 18 | 0+-0 |
| MB-045 | H5-T16311C! | yfull ftdna | C16167T+C16519T | 6 | 12 | 0+-0 |
| MB-046 | H6a1a1a | yfull ftdna | 20 | 26 | 820+-1130 | |
| MB-047 | H6a1a3 | yfull ftdna | non-Ashkenazi in SNP-group A16482G+C16519T | 7 | 113 | 2950+-2660 |
| MB-048 | H6a1a5 | yfull ftdna | A16482G+C16519T | 34 | 53 | 1730+-2320 |
| MB-049 | H6a1b2 | yfull ftdna | insufficient access | 4 | 244 | 4760+-2180 |
| MB-050 | H6a1b3 | yfull ftdna | A16482G | 8 | 93 | 3760+-1850 |
| MB-051 | H7 | yfull ftdna | 573.1C+573.2C+T1700C+T11137C+C152T! | 28 | 191 | 8890+-1680 |
| MB-052 | H7c2 | yfull ftdna | 33 | 35 | 1730+-6570 | |
| MB-053 | H7e | yfull ftdna | 67 | 97 | 1490+-2300 | |
| MB-054 | H11a1 | yfull ftdna | non-Ashkenazi in SNP-group C16519T | 8 | 184 | 2920+-2080 |
| MB-055 | H11a2a | yfull ftdna | 1 | 70 | 3960+-1600 | |
| MB-056 | H11a2a2 | yfull ftdna | G7805A+C16519T+C152T! | 10 | 46 | 1300+-1210 |
| MB-057 | H11b1 | yfull ftdna | C16261T+C152T! | 25 | 62 | 0+-0 |
| MB-058 | H-T152C! | yfull ftdna | T710C+C152T! | 1 | 138 | 0+-0 |
| MB-059 | H10 | yfull ftdna | 1 | 96 | 8590+-2260 | |
| MB-060 | H10a | yfull ftdna | insufficient access | 3 | 53 | 6040+-2360 |
| MB-061 | H10a1b | yfull ftdna | 20 | 22 | 720+-1170 | |
| MB-062 | H10e | yfull ftdna | 1 | 207 | 4680+-2260 | |
| MB-063 | H13a1a1 | yfull ftdna | insufficient access | 3 | 71 | 5880+-1890 |
| MB-064 | H13a1a1a | yfull ftdna | 1 | 108 | 4490+-2180 | |
| MB-065 | H15b | yfull ftdna | C16519T+C16527T | 7 | 25 | 5500+-3300 |
| MB-066 | H17a | yfull ftdna | 1 | 41 | 6090+-2690 | |
| MB-067 | H23 | yfull ftdna | 1 | 178 | 2060+-2840 | |
| MB-068 | H25 | yfull ftdna | 12 | 12 | 0+-0 | |
| MB-069 | H26c | yfull ftdna | 31 | 34 | 2940+-7620 | |
| MB-070 | H40b | yfull ftdna | 41 | 44 | 1110+-6020 | |
| MB-071 | H41a | yfull ftdna | C14433T+C16519T | 18 | 79 | 0+-0 |
| MB-072 | H44a1 | yfull ftdna | 1 | 31 | 0+-0 | |
| MB-073 | H47 | yfull ftdna | insufficient access | 3 | 32 | 8930+-3030 |
| MB-074 | H56 | yfull ftdna | non-Ashkenazi in SNP-group - | 12 | 88 | 4690+-2980 |
| MB-075 | H65a | yfull ftdna | non-Ashkenazi in SNP-group - | 15 | 30 | 0+-0 |
| MB-076 | HV0-T195C! | yfull ftdna | C7229T+G11963A+C16519T | 5 | 289 | 0+-0 |
| MB-077 | HV1a'b'c | yfull ftdna | G709A+A13098G+T16368C+C16519T+A16129G! | 10 | 37 | 0+-0 |
| MB-078 | HV1b | yfull ftdna | 1 | 9 | 14800+-3690 | |
| MB-079 | HV1b2 | yfull ftdna | 139 | 157 | 1510+-4290 | |
| MB-080 | HV5 | yfull ftdna | insufficient access | 4 | 1 | 2010+-14530 |
| MB-081 | HV5a | yfull ftdna | 66 | 75 | 0+-0 | |
| MB-082 | HV10 | yfull ftdna | 1 | 15 | 2010+-8600 | |
| MB-083 | I1b | yfull ftdna | 1 | 51 | 11130+-4810 | |
| MB-084 | I1c | yfull ftdna | 7 | 8 | 8210+-3780 | |
| MB-085 | I1c1a | yfull ftdna | 44 | 57 | 0+-0 | |
| MB-086 | I2a | yfull ftdna | 1 | 35 | 3770+-2140 | |
| MB-087 | I5a1b | yfull ftdna | 1 | 4 | 0+-0 | |
| MB-088 | I5c1 | yfull ftdna | 1 | 13 | 0+-0 | |
| MB-089 | J1b1a1 | yfull ftdna | C2322T+C16519T | 43 | 275 | 6970+-2340 |
| MB-090 | J1b2 | yfull ftdna | 1 | 102 | 6160+-5560 | |
| MB-091 | J1c1 | yfull ftdna | G228+C7966T+T9497C+G14198A+A14332G+T16093C+C16519T+C16189T!+C16311T! | 32 | 144 | 10090+-2220 |
| MB-092 | J1c1e | yfull ftdna | 1 | 15 | 0+-0 | |
| MB-093 | J1c3e2 | yfull ftdna | insufficient access G185A+G228A+C2280T+A13803G+C15853T+A16235G+C16519T+C16311T! | 5 | 53 | 4910+-3010 |
| MB-094 | J1c4 | yfull ftdna | 1 | 119 | 4070+-3080 | |
| MB-095 | J1c5 | yfull ftdna | 1 | 229 | 10630+-2320 | |
| MB-096 | J1c-C16261T | yfull ftdna | insufficient access G185A+G228A+T16093C+C16519T+G16274A! | 7 | 36 | 0+-0 |
| MB-097 | J1c7 | yfull ftdna | insufficient access | 6 | 53 | 5940+-3960 |
| MB-098 | J1c7a | yfull ftdna | G185A+G228A+T16092C+C16519T | 80 | 231 | 1980+-5370 |
| MB-099 | J1c13 | yfull ftdna | 12 | 16 | 1080+-2240 | |
| MB-100 | J1c14 | yfull ftdna | 50 | 74 | 990+-2910 | |
| MB-101 | J2b1a2a | yfull ftdna | 1 | 12 | 0+-0 | |
| MB-102 | J2b1e | yfull ftdna | 8 | 9 | 0+-0 | |
| MB-103 | K1a1b1 | yfull ftdna | 1 | 77 | 14210+-3010 | |
| MB-104 | K1a1b1a | yfull ftdna | 723 | 752 | 4820+-3570 | |
| MB-105 | K1a4 | yfull ftdna | 2 | 61 | 15940+-2340 | |
| MB-106 | K1a4a | yfull ftdna | C6134T+G6480A+A6605G+C16168T | 7 | 27 | 14090+-2670 |
| MB-107 | K1a5b | yfull ftdna | 1 | 12 | 0+-0 | |
| MB-108 | K1a9 | yfull ftdna | 214 | 227 | 1690+-2370 | |
| MB-109 | K1a11 | yfull ftdna | 1 | 70 | 4760+-2850 | |
| MB-110 | K2a | yfull ftdna | non-Ashkenazi in SNP-group - | 13 | 163 | 9100+-2270 |
| MB-111 | K2a2a | yfull ftdna | incorrect data | 12 | 0 | 1820+-1930 |
| MB-112 | K2a2a1 | yfull ftdna | 185 | 234 | 0+-0 | |
| MB-113 | L2a1-T16189C! | yfull ftdna | 2 | 2 | 0+-0 | |
| MB-114 | L2a1l2a+L2a1l2a1 | yfull ftdna | 77 | 85 | 2370+-2140 | |
| MB-116 | M1a1b1c | yfull ftdna | A16182c of A16183c | 11 | 17 | 0+-0 |
| MB-117 | M33-T16362C | yfull ftdna | 1 | 0 | 0+-0 | |
| MB-118 | M33c | yfull ftdna | C4182T+C4577T+A7364G+A16235G | 12 | 25 | 12760+-6950 |
| MB-119 | N1b1a | yfull ftdna | 1 | 32 | 8980+-4730 | |
| MB-120 | N1b1a2 | yfull ftdna | T4619C+G6249A+T7283C+T13608C+A11914G!+C9335T! | 3 | 46 | 0+-0 |
| MB-121 | N1b1b1 | yfull ftdna | 210 | 246 | 9720+-3660 | |
| MB-122 | N1b2 | yfull ftdna | 2 | 3 | 1180+-2030 | |
| MB-123 | N9a3 | yfull ftdna | 18 | 22 | 8280+-5120 | |
| MB-124 | R0a2c | yfull ftdna | 1 | 27 | 10540+-6060 | |
| MB-125 | R0a2m | yfull ftdna | T58C | 43 | 39 | 4080+-5630 |
| MB-126 | R0a4 | yfull ftdna | 45 | 35 | 1830+-2560 | |
| MB-127 | T1a | yfull ftdna | C6656T | 4 | 65 | 14680+-4820 |
| MB-128 | T1a1 | yfull ftdna | non-Ashkenazi in SNP-group - | 12 | 757 | 6990+-2080 |
| MB-129 | T1a1b | yfull ftdna | G16000A | 3 | 83 | 3860+-2460 |
| MB-130 | T1a1j | yfull ftdna | - | 15 | 29 | 5220+-2490 |
| MB-131 | T1a1k1 | yfull ftdna | 15 | 20 | 0+-0 | |
| MB-132 | T1a5 | yfull ftdna | 1 | 31 | 10840+-5420 | |
| MB-133 | T1b | yfull ftdna | insufficient access | 4 | 37 | 6160+-4800 |
| MB-134 | T1b3 | yfull ftdna | 18 | 19 | 0+-0 | |
| MB-135 | T2a1 | yfull ftdna | non-Ashkenazi in SNP-group T16209C+C16296T+G10398A | 5 | 29 | 15220+-2740 |
| MB-136 | T2a1a8 | yfull ftdna | 1 | 46 | 0+-0 | |
| MB-137 | T2a1b | yfull ftdna | A1530G+A10005G+A11020G+T12481a+T16093C+C16296T | 11 | 31 | 13930+-2680 |
| MB-138 | T2b | yfull ftdna | 1 | 803 | 10060+-1610 | |
| MB-139 | T2b3-C151T | yfull ftdna | 1 | 115 | 0+-0 | |
| MB-140 | T2b4 | yfull ftdna | non-Ashkenazi in SNP-group A16241G | 4 | 166 | 5810+-4850 |
| MB-141 | T2b4a | yfull ftdna | 2 | 80 | 3570+-2840 | |
| MB-142 | T2b-T16362C | yfull ftdna | 1 | 50 | 0+-0 | |
| MB-143 | T2b16 | yfull ftdna | insufficient access | 5 | 21 | 1930+-3690 |
| MB-144 | T2b25 | yfull ftdna | C16296T+A16129G! | 35 | 55 | 6850+-3260 |
| MB-145 | T2e | yfull ftdna | 1 | 108 | 11850+-3230 | |
| MB-146 | T2e1 | yfull ftdna | 1 | 86 | 9170+-2890 | |
| MB-147 | T2e1b+T2e1b1 | yfull ftdna | 18 | 22 | 0+-0 | |
| MB-149 | T2g | yfull ftdna | 1 | 11 | 15710+-5300 | |
| MB-150 | T2g1 | yfull ftdna | 1 | 3 | 3130+-8050 | |
| MB-151 | T2g1a | yfull ftdna | C16296T+C195T! | 3 | 24 | 0+-0 |
| MB-152 | U1b1 | yfull ftdna | A3426G+G5460A+T6365C+G11969A+C16104T+C16111T+A14070G! | 28 | 37 | 11570+-4980 |
| MB-153 | U5a1a1 | yfull ftdna | 1 | 359 | 6830+-3920 | |
| MB-154 | U5a1a1-T152C! | yfull ftdna | 1 | 72 | 0+-0 | |
| MB-155 | U5a1a2a | yfull ftdna | 2 | 57 | 2830+-2530 | |
| MB-156 | U5a1b | yfull ftdna | 2 | 116 | 8370+-2760 | |
| MB-157 | U5a1b1 | yfull ftdna | T15893C+C16519T+C16192T! | 6 | 154 | 6980+-1960 |
| MB-158 | U5a1b1c2 | yfull ftdna | 2 | 7 | 2460+-2210 | |
| MB-159 | U5a1f1a | yfull ftdna | 13 | 14 | 6500+-3590 | |
| MB-160 | U5a2 | yfull ftdna | 1 | 3 | 18410+-3870 | |
| MB-161 | U5a2a1-T152C! | yfull ftdna | 1 | 53 | 0+-0 | |
| MB-162 | U5a2b | yfull ftdna | 1 | 166 | 11430+-3180 | |
| MB-163 | U5a2b2a | yfull ftdna | G15930A+A16265c+C16519T | 4 | 13 | 0+-0 |
| MB-164 | U5b1 | yfull ftdna | 1 | 33 | 15520+-4880 | |
| MB-165 | U5b1b1-T16192C! | yfull ftdna | 2 | 132 | 0+-0 | |
| MB-166 | U5b1e1 | yfull ftdna | 2 | 110 | 2600+-1870 | |
| MB-167 | U5b2 | yfull ftdna | 1 | 21 | 20040+-3200 | |
| MB-168 | U5b2a1a | yfull ftdna | 1 | 22 | 10610+-3810 | |
| MB-169 | U5b2b4a | yfull ftdna | 1 | 80 | 0+-0 | |
| MB-170 | U6a | yfull ftdna | incorrect data | 3 | 0 | 25080+-3740 |
| MB-171 | U6a7a1b | yfull ftdna | C16519T | 12 | 20 | 0+-0 |
| MB-172 | U2e1a1 | yfull ftdna | A8014G+G13708A+A16183c | 10 | 157 | 6150+-2590 |
| MB-173 | U3a1 | yfull ftdna | insufficient access | 5 | 105 | 8910+-3840 |
| MB-174 | U3a1a | yfull ftdna | A257G | 15 | 65 | 6840+-3820 |
| MB-175 | U3a1c1 | yfull ftdna | 1 | 59 | 0+-0 | |
| MB-176 | U3b1b | yfull ftdna | 1 | 39 | 1890+-2620 | |
| MB-177 | U4a | yfull ftdna | 1 | 304 | 14940+-3340 | |
| MB-178 | U4a3 | yfull ftdna | 1 | 3 | 4140+-2780 | |
| MB-179 | U4a3a | yfull ftdna | T13659C | 7 | 62 | 0+-0 |
| MB-180 | U4b1b1 | yfull ftdna | 1 | 103 | 6350+-3290 | |
| MB-181 | U4c1 | yfull ftdna | 1 | 160 | 2280+-2180 | |
| MB-182 | U7a | yfull ftdna | 54 | 68 | 16710+-3010 | |
| MB-183 | U8b1b1 | yfull ftdna | 2 | 37 | 1830+-2750 | |
| MB-184 | V1a1 | yfull ftdna | non-Ashkenazi in SNP-group C16519T | 21 | 149 | 5500+-2160 |
| MB-185 | V7a | yfull ftdna | T89C+G930A+A12753G+C16519T+C195T! | 84 | 127 | 6020+-2410 |
| MB-186 | V7b | yfull ftdna | C16173T,C16519T | 14 | 30 | 1910+-1820 |
| MB-187 | V15 | yfull ftdna | T16093C+T16209C+C16519T | 6 | 10 | 6470+-3270 |
| MB-188 | V18a | yfull ftdna | 2 | 30 | 0+-0 | |
| MB-189 | V21 | yfull ftdna | 1 | 13 | 1160+-7470 | |
| MB-190 | W1h | yfull ftdna | A4917G+A5582G+A16265G+C16292T! | 14 | 22 | 0+-0 |
| MB-191 | W3a1a1 | yfull ftdna | 30 | 29 | 9600+-2610 | |
| MB-192 | W3b1 | yfull ftdna | 33 | 34 | 1510+-2090 | |
| MB-193 | W6a | yfull ftdna | 1 | 59 | 6440+-4170 | |
| MB-194 | X2b7 | yfull ftdna | C198T+A10634G+T14110C+A16183c. C146T! | 25 | 43 | 6550+-3910 |
| MB-195 | X2e2 | yfull ftdna | incorrect data | 1 | 0 | 9740+-5150 |
| MB-196 | X2e2a | yfull ftdna | insufficient access | 5 | 27 | 6050+-5150 |
| MB-301 | J2b1f | yfull ftdna | Mountain Jews 5899.1C,C10223T,C16519T+A11914G! | 13 | 0 | |
| MB-302 | H13a2b1 | yfull ftdna | Mountain and Baghdad Jews 5899.1C (partly C16519T) | 8 | 0 | |
| MB-303 | H14a | yfull ftdna | Baghdad Jews A189G,G13194A,T16249C,C16519T,C16311T! | 2 | 0 | |
| MB-304 | U7a4a1 | yfull ftdna | Mountain Jews C13984T,A16309G | 4 | 0 | |
| MB-305 | T2g1a1 | yfull ftdna | Mountain Jews A4012G,A4562G | 2 | 0 | |
| MB-401 | H2a2a1 | yfull ftdna | Moroccan Jews T5557C,G5585A,T6044C,T8277C,G10373A,C16519T | 4 | 0 | |
| MB-402 | H1e8 | yfull ftdna | Moroccan Jews | 8 | 0 | |
| MB-403 | HV0d | yfull ftdna | Moroccan Jews C4904A (and others) | 6 | 0 | |
| MB-404 | J2a1a1 | yfull ftdna | Moroccan Jews T2392C | 4 | 0 | |
| MB-405 | HV1a2 | yfull ftdna | Moroccan Jews (insufficient access) | 3 | 0 | |
| MB-406 | K1c2 | yfull ftdna | Moroccan Jews C3738T,A8146G | 2 | 0 | |
| MB-407 | H3c1 | yfull ftdna | Tunisian Jews (and others) A189G,C16260T,C16189T! | 2 | 0 | |
| MB-306 | U7a3a | yfull ftdna | Iraq Jews T8167C | 2 | 0 | |
| MB-307 | T2c1a1 | yfull ftdna | Iraq and Baghdad Jews C6998T | 10 | 0 |