






 |  |  |  |  |  | 
|
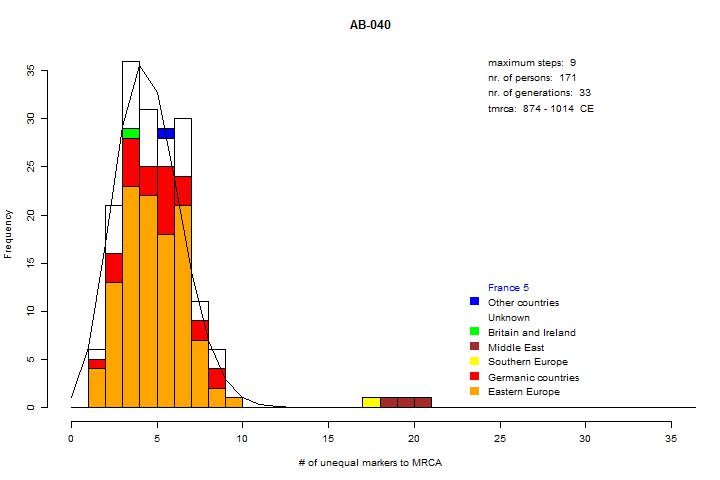
This group has been studied several times. A nice collection can be found in Wirth ftdna site. The distribution in the diagram below suggests a Middle East-Sephardic line.
On the origin on CTS2906: as indicated on http://tree.j2-m172.info/?Hg=J2a1a1b1a1 four lines are known below CTS2906. One is from the Sardinian paper (Francalacci et al.), one from Toscane from the 1kG project, One has not reported an origin (311816), and one is the Ashkenazi branch. The length is on average 45 FGC markers, so about 4000 ybp. In the ftdna project we have a person from the Netherlands and one from Italy. Where and when a person became Jewish is unknown. Two markers were shared between the Ashkenazi branch and the person from unreported origin. Since the BAM files of the other two branches are not analysed for these markers, we do not know if they share the same markers. Near CTS2906 several branches were found in the Toscani region from the 1kG and Sardinian project. It is more likely that this group increased in size in Italy or neighboring area after the shared ancestor lived there, than that the individuals arrived as a group from one location a thousand years later to this area.
In the ftdna project site Jewish DNA we can find E11451 (Marquez) and 271540 (Turkey) at a distance of about 1000 years, suggesting that some members left Iberia after the Inquisition. This is a very small percentage.
Genealogical study: A study by the Bacharach family (from Frankfurt DNA project) is found on http://bacharachdna.com/. In the beginning of the 16th century four German cities had many Jews: Prague was the largest, followed by Vienna, Frankfurt and Worms.
On the Jews of Frankfurt website a very nice Y-DNA STR familytree is shown which follows the historic information of the movements of the family members.
On the 2013 IAJGS Conference on Jewish Genealogy in Boston a presentation and paper was presented on the genealogical data and Y-DNA analysis of this branch. Many of the familynames in the first period in Germany followed the names of the cities: Bacharach and Treves in the Rhein-Frankfurt region, and later Fürth, Wertheim, Weil (see also Weil surname), and Fürth in the direction of Bavaria. The Jewish history of these cities in these periods (1000-1300 CE) is well known. The origin of other names like Schwab could be related, but is more uncertain.
On the STR and SNP variability in this Ashkenazi branch: this branch has several STR characteristics that are grouped. We can compare the subranches of Treves-Weill with Bacharach-Wertheimer, because of the Wirth project diagram. The Bacharach branch has DYS464=13-13-16-16-16-16 (Treves: 13-13-15-16-16-16), DYS557=14 (Treves: 15), DYS446=often 14 (Treves: 13), DYS505=13 (Treves: 14), DYS549=12 (Treves: 13), DYS463=22 (Treves: 21). This suggests that males were 6 mutations took place between Bacharach-Wertheimer and Treves-Weill. If we determine the tmrca of the two ancestors, they are 1240 and 1150. This means that we have two scenarios, which can be checked with next generation SNP analysis: the two are parallel relatives, or MRCA of the first groups is a descendant of the MRCA of the second group; in this case the first ancestor had a somewhat larger population growth than the second. Because of the similar tmrca of both groups, it seems more likely that they were parallel. The shared ancestor would be a little earlier, like 1000 ybp.
You can find additional information in this ftdna project .
You can find additional information in this Ashkenazi branch of ftdna by Jeffrey D. Wexler in 2019 .
This branch is in the J2a-group of Jewish Branches.
This branch is a member of the Ashkenazi branches
| group | Name | Ancestor | Country | SNP | D Y S 3 9 3 | D Y S 3 9 0 | D Y S 1 9 | D Y S 3 9 1 | D Y S 3 8 5 | D Y S 4 2 6 | D Y S 3 8 8 | D Y S 4 3 9 | D Y S 3 8 9 i | D Y S 3 9 2 | D Y S 3 8 9 i i | D Y S 4 5 8 | D Y S 4 5 9 | D Y S 4 5 5 | D Y S 4 5 4 | D Y S 4 4 7 | D Y S 4 3 7 | D Y S 4 4 8 | D Y S 4 4 9 | D Y S 4 6 4 | D Y S 4 6 0 | Y - G A T A - H 4 | Y C A I I | D Y S 4 5 6 | D Y S 6 0 7 | D Y S 5 7 6 | D Y S 5 7 0 | C D Y | D Y S 4 4 2 | D Y S 4 3 8 | D Y S 5 3 1 | D Y S 5 7 8 | D Y F 3 9 5 S 1 | D Y S 5 9 0 | D Y S 5 3 7 | D Y S 6 4 1 | D Y S 4 7 2 | D Y F 4 0 6 S 1 | D Y S 5 1 1 | D Y S 4 2 5 | D Y S 4 1 3 | D Y S 5 5 7 | D Y S 5 9 4 | D Y S 4 3 6 | D Y S 4 9 0 | D Y S 5 3 4 | D Y S 4 5 0 | D Y S 4 4 4 | D Y S 4 8 1 | D Y S 5 2 0 | D Y S 4 4 6 | D Y S 6 1 7 | D Y S 5 6 8 | D Y S 4 8 7 | D Y S 5 7 2 | D Y S 6 4 0 | D Y S 4 9 2 | D Y S 5 6 5 | D Y S 7 1 0 | D Y S 4 8 5 | D Y S 6 3 2 | D Y S 4 9 5 | D Y S 5 4 0 | D Y S 7 1 4 | D Y S 7 1 6 | D Y S 7 1 7 | D Y S 5 0 5 | D Y S 5 5 6 | D Y S 5 4 9 | D Y S 5 8 9 | D Y S 5 2 2 | D Y S 4 9 4 | D Y S 5 3 3 | D Y S 6 3 6 | D Y S 5 7 5 | D Y S 6 3 8 | D Y S 4 6 2 | D Y S 4 5 2 | D Y S 4 4 5 | Y - G A T A - A 1 0 | D Y S 4 6 3 | D Y S 4 4 1 | Y - G G A A T - 1 B 0 7 | D Y S 5 2 5 | D Y S 7 1 2 | D Y S 5 9 3 | D Y S 6 5 0 | D Y S 5 3 2 | D Y S 7 1 5 | D Y S 5 0 4 | D Y S 5 1 3 | D Y S 5 6 1 | D Y S 5 5 2 | D Y S 7 2 6 | D Y S 6 3 5 | D Y S 5 8 7 | D Y S 6 4 3 | D Y S 4 9 7 | D Y S 5 1 0 | D Y S 4 3 4 | D Y S 4 6 1 | D Y S 4 3 5 |
|
| AB-040 | J2a-M92-L556 | 12 | 22 | 14 | 10 | 14-15 | 11 | 15 | 12 | 14 | 11 | 31 | 15 | 9-9 | 11 | 11 | 26 | 15 | 20 | 29 | 13-13-15-16-16-16 | 10 | 10 | 19-22 | 15 | 12 | 15 | 17 | 34-36 | 12 | 9 | 10 | 7 | 15-15 | 8 | 11 | 10 | 8 | 11 | 9 | 12 | 17-17 | 15 | 10 | 12 | 12 | 15 | 8 | 11 | 22 | 21 | 13 | 12 | 11 | 13 | 11 | 12 | 12 | 11 |
List of SNPs in yfull tree:Home - A0-T - A1 - A1b - BT - CT - CF - F - GHIJK - HIJK - IJK - IJ - J - J2 - M410 - PF4610 - L26 - PF5087 - PF5116 - PF5119 - CTS4800 - L558 - M67 - Z1847 - Z500 - M92 - Z508 - Z504 - Z8096 - Y20492 - Y20051 - L560 - L556
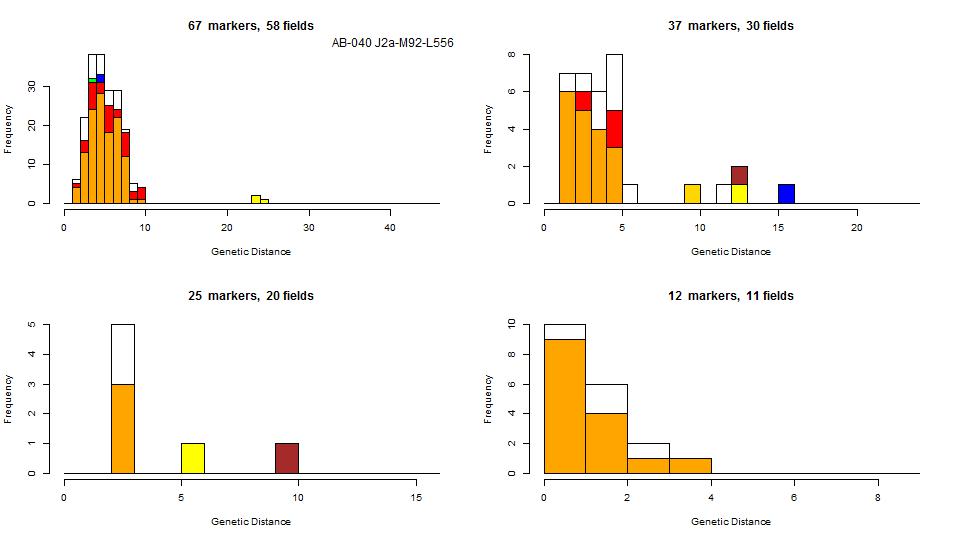
The following kitnr's are recognized in this group:
N29103
71691
70610
119546
8416
101015
211627
86120
99705
29072
N36595
15854
78928
58014
167079
50626
228035
46571
101722
108965
N22878
101459
150891
424
30979
222781
260202
105493
208921
916
194828
163
116289
181727
49024
47749
247255
298374
N41195
157655
N26932
175667
150344
343
43230
103387
5984
101977
134363
N56522
N54227
157062
N78385
411
8415
19438
N80760
199842
65940
297320
313259
232883
3205
188569
36358
61810
49388
132750
161421
176782
N24922
211622
1
302142
116068
147963
8811
67387
6034
150661
73760
61746
242488
22025
69509
122839
209229
254537
70867
172784
163184
73647
259528
3206
70612
262029
223292
113336
N31200
148429
28924
156873
145711
200675
189752
28730
162907
167053
153990
163122
N2500
188861
97190
240551
N29430
162966
174705
222819
230191
N27113
N69241
23075
50911
175250
E12266
19386
137126
197945
141290
170586
68220
145757
118449
186146
168104
137121
112842
196489
20908
204432
163407
296689
174146
284980
46925
73944
118655
55452
N41658
72245
282535
104201
112303
216297
171079
114078
178971
275611
312867
163611
195735
168314
187786
158753
N79686
149535
238369
N31492
226303
221907
197833
Kits with nearby 67-markers are found in semargl.me
Check for yourself by inspecting the data that was used.